3 S3 vs S4
The lm object that we saw in the previous example is an object and it uses the S3 object system. The other built-in object system is called S4, which is used by the mixed-effect modeling package lme4 and the Bioconductor packages (common packages: limma and DESeq2 and others for genomic analyses).
3.1 S3
S3 is used by the most common R modeling methods like lm() and glm(). You access elements of the objects using the dollar-sign, like my_model$coefficients.
The S3 object system is very loosey-goosey. This happened because R wasn’t developed with the intention of having an object system, so S3 was pieced together from parts that were already there in the language, but intended for other purposes. Here is how easy it is to create a whole new S3 class and an object of the class, including members and methods:
my_obj = list()
my_obj$a = 1:4
my_obj$b = 10:7
# turn my_obj into an object by assigning it a class name
class(my_obj) = "dumb"
# define a plot method for the class
plot.dumb = function( obj ) plot(obj$a, obj$b)
plot(my_obj)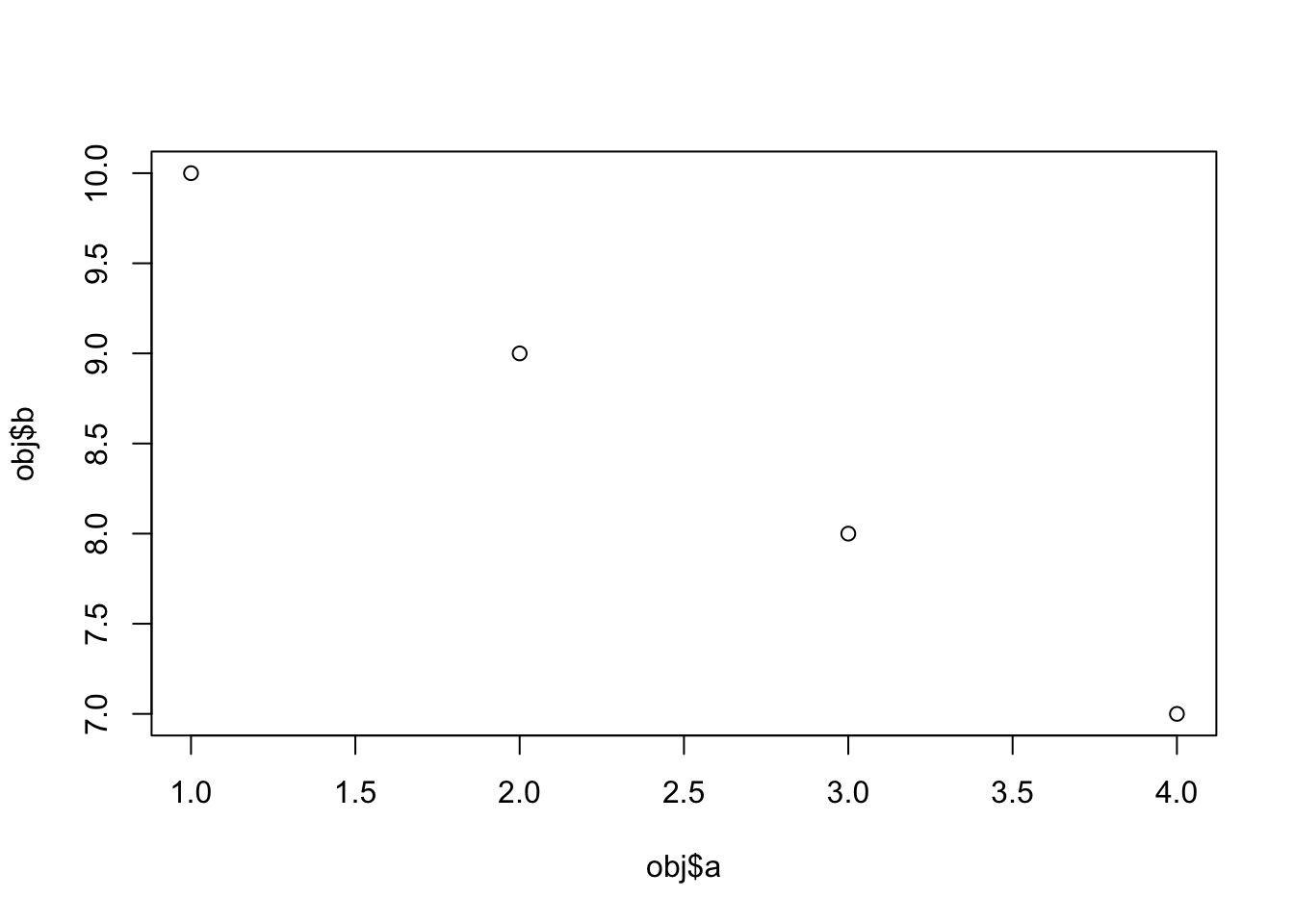
This flexibility is very liberating, but can also cause problems. There is nothing that enforces the expectation that object of class dumb must have members a and b, so you can crash the plot() method:
obj2 = list()
obj2$message = "No bloody a, b, or c"
# turn my_obj into an object by assigning it a class name
class(obj2) = "dumb"
# define a plot method for the class
plot.dumb = function( obj ) plot(obj$a, obj$b)
plot(obj2)Warning in min(x): no non-missing arguments to min; returning InfWarning in max(x): no non-missing arguments to max; returning -InfWarning in min(x): no non-missing arguments to min; returning InfWarning in max(x): no non-missing arguments to max; returning -InfError in plot.window(...): need finite 'xlim' values
The S4 object system was developed in order to impose some order to the chaos.
##S4 Used by lme4 and Bioconductor. The S4 object system is much more formal than S3 - a class must be fully defined before it can be used, and objects can only be modified through their methods. We aren’t going to create a new S4 class because it is much more complicated that the S3 example. Instead, let’s generate a mixed-effects model using the lmer() function from the lme4 package.
install.packages( "lme4" )
The downloaded binary packages are in
/var/folders/7x/s3x3ymxs1_lcppnqy99_4kx40000gp/T//RtmpfwJWvr/downloaded_packageslibrary(lme4)# estimate the linear, mixed-effects model:
peng_lme = lmer( body_mass_g ~ species + sex + (1|island), data=penguins )boundary (singular) fit: see help('isSingular')The easiest way to know that peng_lme is an S4 object is that it is listed as a “Formal class” in the “Environment” pane of RStudio.
The next most visible difference is that you can’t use $ to access the members that are encapsulated by an S4 class. Instead, you use @ or the slot() function:
# inspect the slot "beta"
peng_lme@beta[1] 3372.38681 26.92385 1377.85803 667.55515# the preferred way to do the same:
slot(peng_lme, "beta")[1] 3372.38681 26.92385 1377.85803 667.55515# You get an error if you try using $ to access a slot:
peng_lme$betaError in peng_lme$beta: $ operator not defined for this S4 classAnother thing you may notice is that you can’t change the structure of an S4 object:
# get an error if you try to conjure a new slot:
peng_lme@test = 5Error in (function (cl, name, valueClass) : 'test' is not a slot in class "lmerMod"